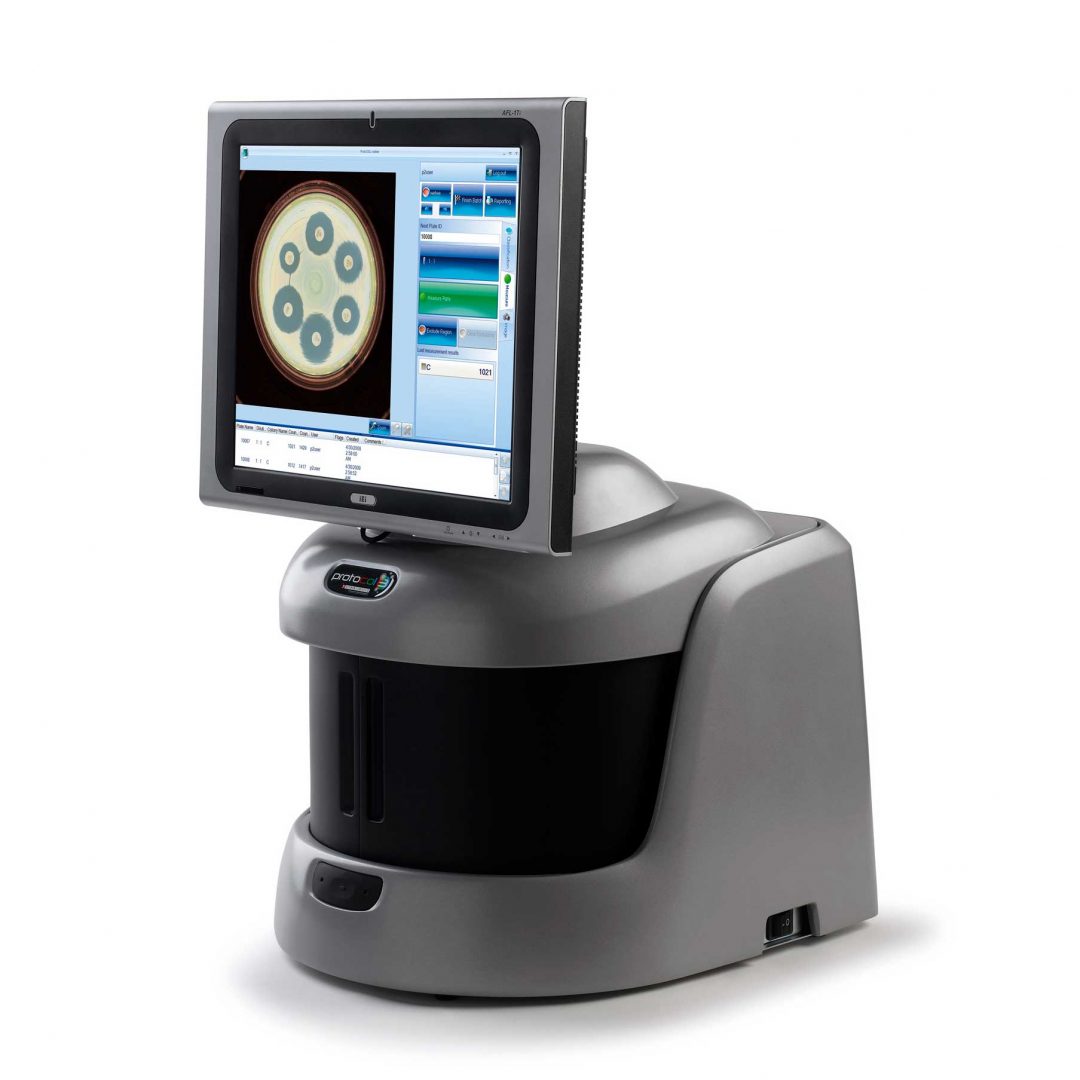
Figure 1. Images of pour plate before and after analysis in ProtoCOL 3
Colony counting
Bacterial colony counting is widely used in both industry and research laboratories in a wide range of applications; these include quality control, environmental monitoring, immunological studies and medical testing. The number of colonies on an agar plate can be used to estimate the number of viable bacteria (total viable count) present in a test sample. This can then be used as an indicator of the cleanliness of a surface, the sterility of a product or the presence of a bacterial infection.
Traditionally, colony counting was performed manually or using a light box which was time-consuming and prone to human error. The advent of automated colony counters, which use sophisticated algorithms to detect and count colonies based on shape or colour, has overcome these drawbacks.
The ‘Pour Plate’ module comes as standard in the ProtoCOL 3, Protos 3, and aCOLyte 3 HD software and allows colony counting in a variety of different formats. Plate names can be entered manually, configured in a plate list, transferred through LIMS or read by a barcode reader. The module can analyse circular, rectangular and even partial plate areas. Colonies as small as 43 µm can be detected based on their contrast against the background/media or based on their colour, shape or size. This allows the Pour Plate module to accommodate a wide range of different bacterial morphologies and agar types.
The sensitivity of colony detection can be altered depending on the type of plate being analysed and debris can be removed by enabling the ‘reject small particles’ function.
The sophisticated software is also capable of splitting touching colonies, ensuring that counts are accurate even on plates with a high bacterial density. The software can detect and remove visible grids, making it easier to count colonies on gridded membranes and water plates.
Dilutions can be entered in the software which produces accurate count/ml results. Minimum and maximum count restrictions can also be set to produce </> results for out of spec counts. Accuracy and small particle rejection limits can be adjusted between plates and results can be recorded as ‘zero’ or ‘too numerous to count’ if necessary. Certain functions, such as manual counts and excluded regions are flagged in the audit report for full traceability.
Compliance
All colony count results are automatically transferred into a table for storage in a secure database. The database is password protected and so ensures that batches of results cannot be deleted. Additionally, count editing is recorded with a coded flag next to the revised result. Every detail of the sample including pictures of the plates, system configuration, user details, date and time are recorded in professional reports (Excel, Open Office or PDF). For laboratories that require the highest level of compliance and traceability, the ProtoCOL 3 system comes with a full audit trail, user access levels and LIMS connection.
Synbiosis releases free bi-annual software upgrades which can be easily downloaded from the website. This means that customers have access to any improvements and new features that are added to the software.
Common Applications: Bioburden test, Microbial Limits Test, Total Viable Count, Preservative Efficacy Testing, Environmental Monitoring, Water Testing, Settle Plate, Contact Plate, and Membrane Filtration.
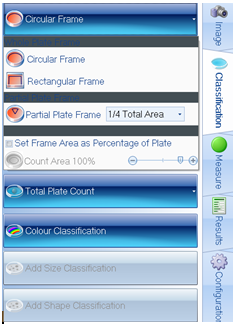
Figure 2. Plate and colony classification tab within ProtoCOL 3 software
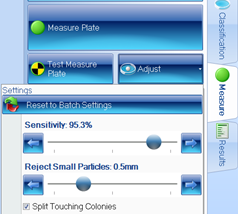
Figure 3. Measure tab within ProtoCOL 3 software showing adjustment window
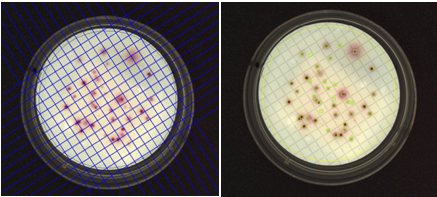
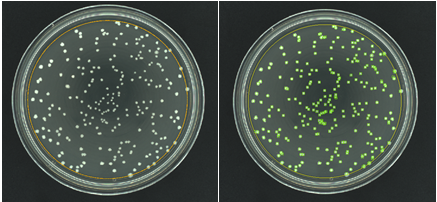
Figure 4. Images of a membrane filtration plate during grid removal (left) and after analysis (right) in the ProtoCOL 3 software (colonies are marked with green squares)
Figure 5. Measure tab within ProtoCOL 3 software showing additional features