Opsonophagocytic Killing Assay (OPKA)
What is an OPK assay?
OPK assays play a key role in developing and improving vaccines, particularly pneumococcal vaccines. They are a useful addition to the pneumococcal antibody enzyme-linked immunosorbent assay (ELISA). The assay is used to quantify opsonic antibody activity present in blood serum samples. This in-vitro assay aids in selecting promising vaccines by demonstrating whether the vaccine-induced antibodies drive efficient complement deposition and subsequent opsonophagocytic killing. OPK assays make it possible to reproducibly estimate the phagocytic titre of sera from vaccinated and un-vaccinated individuals. The antigenic target is viable bacteria. Effective opsonisation mediates phagocytic cell death of the bacteria.
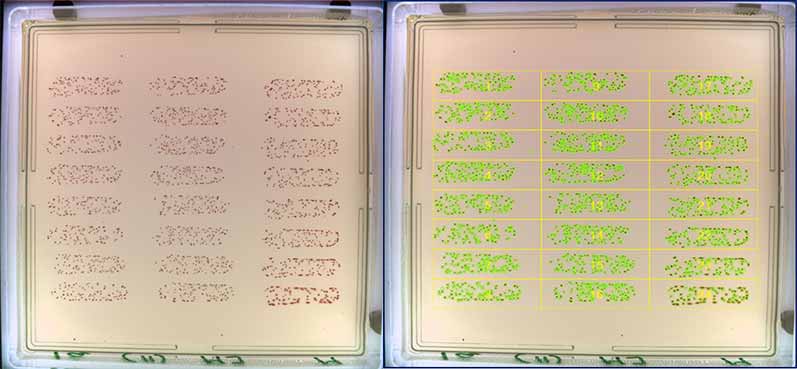
Figure 1. Before and after images of OPKA analysis in the ProtoCOL 3
OPKA method
To carry out an OPK assay, bacterial strains are incubated with sera and complement. Agar plates are then inoculated with the bacterial cultures at different dilutions and incubated overnight. The number of colonies present on the agar can be compared to controls and used to assess the percentage bacterial kill of the sera.
Benefits of automation
Since colony enumeration provides the data on which vaccine efficacy is based, it is essential to obtain precise colony counts. Traditionally, manual methods of enumerating the OPK assay required microbiologists to use a light box and pen and then key the results manually into a computer. A study by Putman et al (2005) compared the efficiency of the ProtoCOL to a scanner, digital camera and manual counting. The results demonstrated the ability of the ProtoCOL to overcome many of the hurdles associated with traditional methods of enumeration such as reading time and transcription errors. Additionally, because traditional methods do not produce any digital images of the plate alongside the colony count, automated colony counters improve audit readiness vital for the approval of new vaccines.
ProtoCOL 3 colony counter
The ProtoCOL 3 software enables the scientist to read OPKA plates in a variety of formats from single columns to 12 x 12 grids. The software allows the sensitivity to be altered and small particles and debris to be excluded based on a selected size (mm). The software can also split closely touching colonies, giving a more accurate count in areas with high bacterial density. Colonies can be classified based on their colour, contrast, size or shape. The software allows up to 20 different colours to be detected and can detect colonies as small as 43 µm.
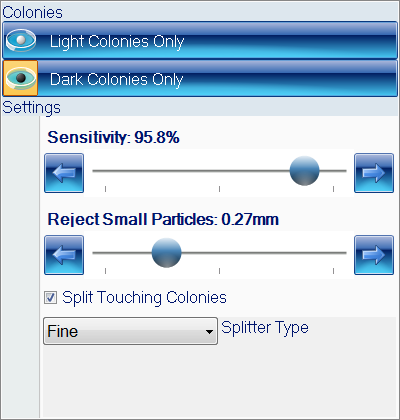
Figure 2. Colony classification step in ProtoCOL 3 OPKA software
The ProtoCOL 3 system is fully CFR 21 Part 11 compliant with a full audit trail. Results produced from OPK assays are compliant with GCP (Good Clinical Practice) guidelines which can be presented to regulatory authorities such as the European Medicines Control Agency (EMEA) and Food and Drug Administration (FDA).
The colony count results are automatically transferred into a table for storage in a secure database. The database is password protected and so ensures that batches of results cannot be deleted. Additionally, count editing is recorded with a coded flag next to the revised result. Every detail of the sample including pictures of the OPKA plates, system configuration, user details, date and time are recorded in a professional report (Excel, Open Office or PDF).
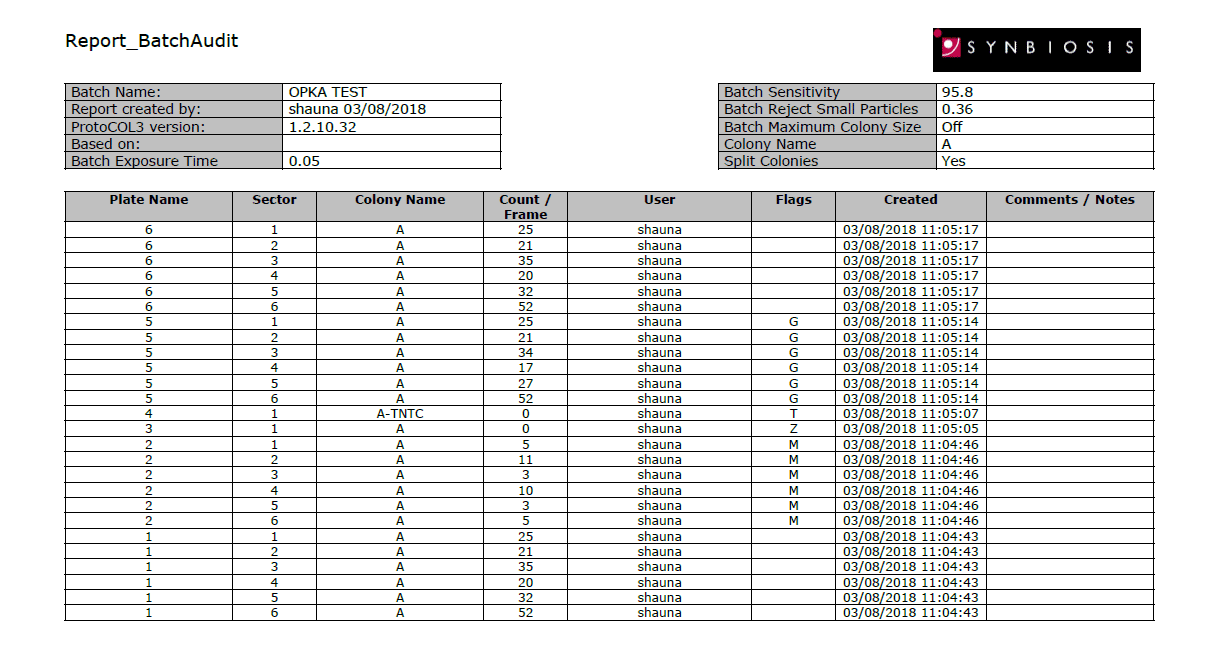
Figure 3. Example of a PDF OPKA report produced in ProtoCOL 3 software.
Putman, M., Burton, R. & Nahm, MH. (2005). Simplified method to automatically count bacterial colony forming unit. Journal of Immunological Methods. 302. 99-102.
