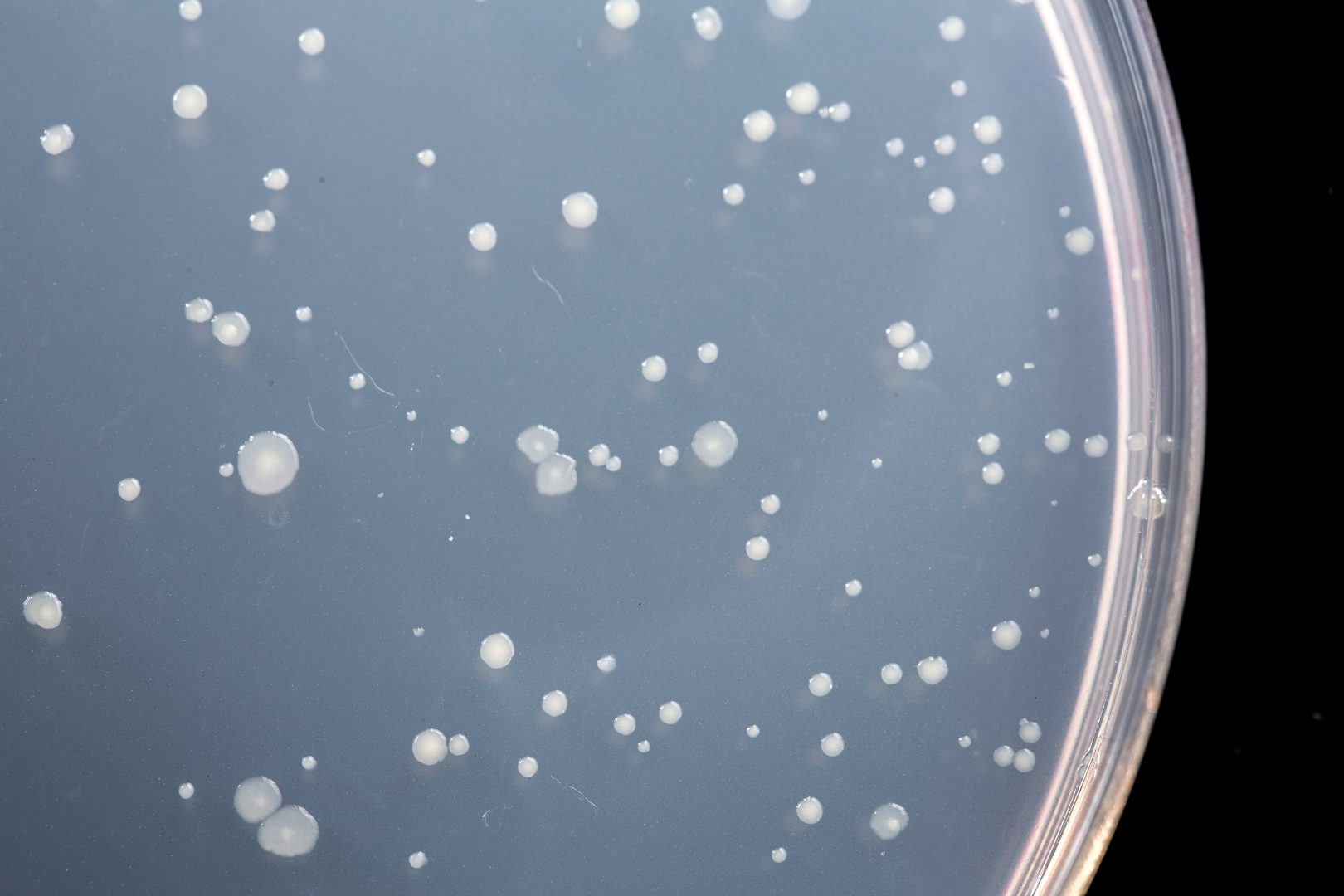
Ames Testing
What is Ames testing?
Ames testing allows researchers to determine the degree of probable mutagenic activity likely to occur in the presence of one or more chemicals. It is a fast and inexpensive method of predicating whether or not a chemical poses a carcinogenic risk from causing mutations in genetic material. Salmonella typhimurium strains that lack the gene to produce histidine are routinely used as the bacterium of choice for the Ames test.
Ames test method
Chemically-treated bacteria are grown on agar plates deficient in the amino acid histidine. Mutated histidine-dependant Salmonella cannot form visible colonies under such conditions unless they undergo a second mutation and revert back to being histidine-producing. The colonies that undergo this mutation in the presence of the chemical of interest can then grow on the agar. These colonies are reported as the ‘numbers of revertant colonies’ per plate based on concentration of test chemical. Ames results are often expressed as a ratio of revertant colonies on test plates/ colonies on negative control.
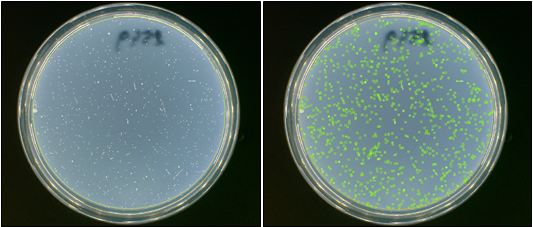
Figure 1. Images of Ames plates before and after analysis in the ProtoCOL 3
Benefits of automation
Since the results of Ames tests are based on the ‘number of revertant colonies’, it is essential to obtain precise colony counts. Manual colony counting methods require microbiologists to use a light box and pen and key the results manually into a computer. This can be time-consuming and can lead to reading and transcription errors, especially when counting large numbers of colonies. Additionally, traditional methods do not produce digital images of the plate alongside the colony count, making independent audits difficult to carry out.
ProtoCOL 3 automated counting
The ProtoCOL 3 software allows researchers to easily and reproducibly count the colonies present on Ames plates. The system enables the user to read both circular and rectangular plates in either a full or partial frame. ProtoCOL 3 will count colonies as small as 43 microns. The software also allows the detection sensitivity to be altered and small particles/debris to be excluded based on a selected size (mm). ProtoCOL 3 provides the option to split touching colonies, giving a more accurate count in areas with high bacterial density. Researchers can choose to enter plate names manually or for plate numbers to increase incrementally for each plate. If the same plate names are regularly used for Ames testing, a plate list can be saved and used to speed up testing. The ProtoCOL 3 also works with barcode scanners and LIMS systems. Dilution factors can be applied to each plate to provide accurate count/plate or count/ml results.
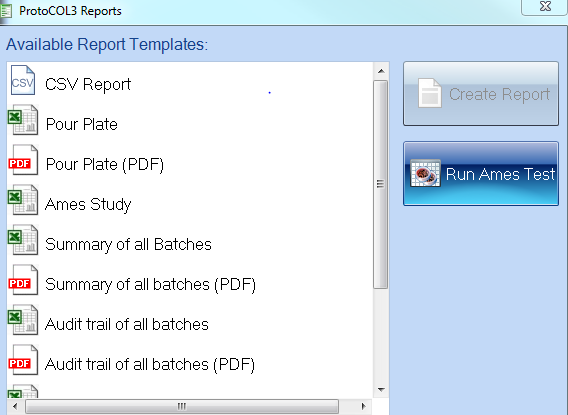
Figure 2. ProtoCOL 3 report options window including Ames Study Manager
Our newly developed ‘Ames Study Manager’ saves valuable time by allowing researchers to analyse their experimental data within ProtoCOL 3. The Study Manager allows details of bacterial strains, chemicals, controls, and replicates to be entered, producing a professional study report. Mean count results are compared to negative ‘solvent controls’ to produce a ratio of probable mutagenic activity in each case.
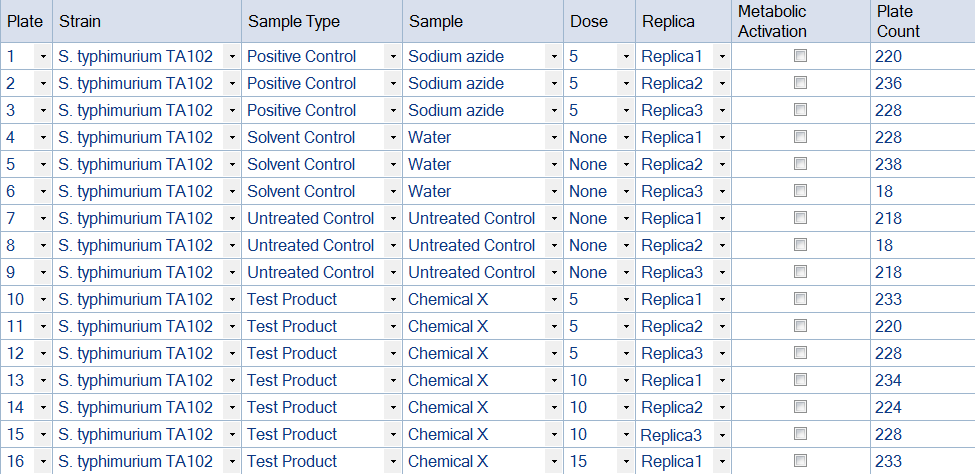
Figure 3. Example of an Ames Study template produced in ProtoCOL 3
ProtoCOL 3 Compliance
The ProtoCOL 3 system is fully ‘CFR 21 Part 11’ compliant with a full audit trail. ProtoCOL 3 produces Ames testing results that are compliant with GLP (Good Laboratory Practice) guidelines which can be presented to regulatory authorities such as the European Medicines Control Agency (EMEA) and Food and Drug Administration (FDA). The colony count results are automatically transferred into a table for storage in a secure database. The database is password protected and so ensures that batches of results cannot be deleted. Additionally, count editing is recorded with a coded flag next to the revised result. Every detail of the sample including pictures of the Ames plates, system configuration, user details, date and time are recorded in professional reports (Excel, Open Office or PDF).
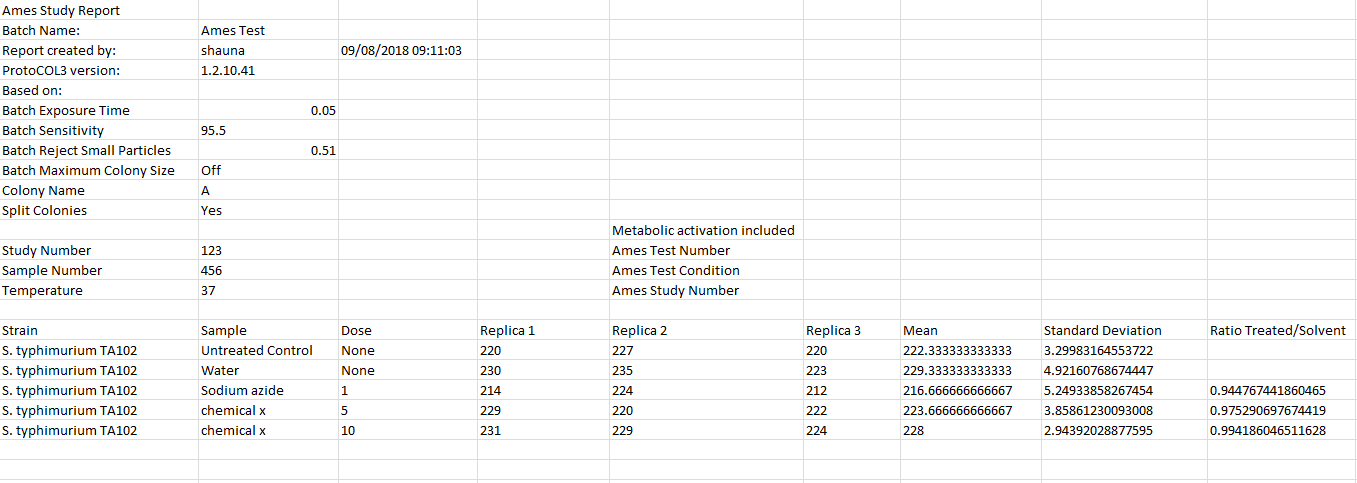
Figure 4. Example of an Ames Study report produced in ProtoCOL 3
